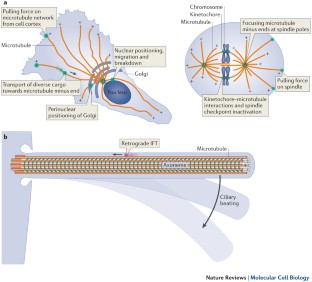
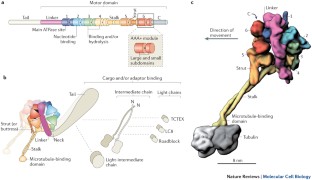
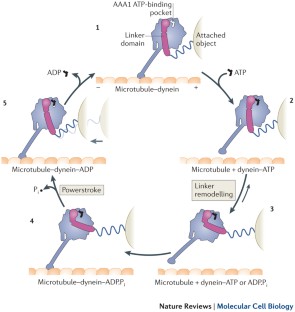
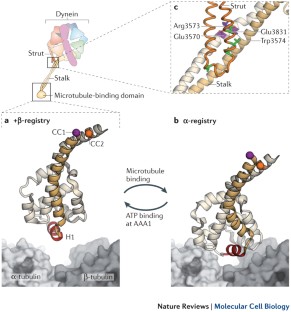
Functions and mechanics of dynein motor proteins
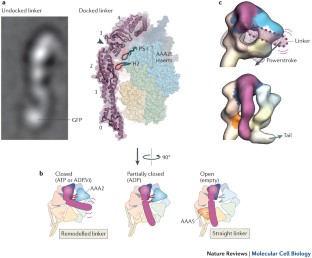
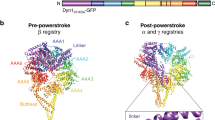
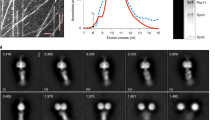
Fuelled by ATP hydrolysis, dyneins generate force and movement on microtubules in a wealth of biological processes, including ciliary beating, cell division and intracellular transport. The large mass and complexity of dynein motors have made elucidating their mechanisms a sizable task. Yet, through a combination of approaches, including X-ray crystallography, cryo-electron microscopy, single-molecule assays and biochemical experiments, important progress has been made towards understanding how these giant motor proteins work. From these studies, a model for the mechanochemical cycle of dynein is emerging, in which nucleotide-driven flexing motions within the AAA+ ring of dynein alter the affinity of its microtubule-binding stalk and reshape its mechanical element to generate movement.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
206,07 € per year
only 17,17 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
The regulatory function of the AAA4 ATPase domain of cytoplasmic dynein
Article Open access 23 November 2020

Nde1 promotes Lis1-mediated activation of dynein
Article Open access 09 November 2023
Pac1/LIS1 stabilizes an uninhibited conformation of dynein to coordinate its localization and activity
Article 27 April 2020
Accession codes
Accessions
Protein Data Bank
References
- Moughamian, A. J. & Holzbaur, E. in Dyneins: Structure, Biology and Disease (ed. King, S. M) 584–601 (Elsevier Inc., 2011).
- Fliegauf, M., Benzing, T. & Omran, H. When cilia go bad: cilia defects and ciliopathies. Nature Rev. Mol. Cell Biol.8, 880–893 (2007). ArticleCASGoogle Scholar
- Gibbons, I. R. Studies on the protein components of cilia from Tetrahymena pyriformis. Proc. Natl Acad. Sci. USA50, 1002–1010 (1963). ArticleCASPubMedPubMed CentralGoogle Scholar
- Gibbons, I. & Rowe, A. Dynein: a protein with adenosine triphosphatase activity from cilia. Science149, 424–426 (1965). References 3 and 4 mark the discovery of dynein motor proteins.ArticleCASPubMedGoogle Scholar
- Paschal, B. M., Shpetner, H. S. & Vallee, R. B. MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J. Cell Biol.105, 1273–1282 (1987). Describes the isolation and characterization of cytoplasmic dynein.ArticleCASPubMedGoogle Scholar
- Paschal, B. M. & Vallee, R. B. Retrograde transport by the microtubule-associated protein MAP 1C. Nature330, 181–183 (1987). ArticleCASPubMedGoogle Scholar
- Schroer, T. A., Steuer, E. R. & Sheetz, M. P. Cytoplasmic dynein is a minus end-directed motor for membranous organelles. Cell56, 937–946 (1989). ArticleCASPubMedGoogle Scholar
- Vale, R. D., Reese, T. S. & Sheetz, M. P. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell42, 39–50 (1985). ArticlePubMedPubMed CentralCASGoogle Scholar
- Vale, R. D. & Milligan, R. A. The way things move: looking under the hood of molecular motor proteins. Science288, 88–95 (2000). ArticleCASPubMedGoogle Scholar
- Billington, N. & Sellers, J. R. Dynein struts its stuff. Nature Struct. Mol. Biol.18, 635–636 (2011). ArticleCASGoogle Scholar
- Neuwald, A. F., Aravind, L., Spouge, J. L. & Koonin, E. V. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res.9, 27–43 (1999). CASPubMedGoogle Scholar
- Vallee, R. B., McKenney, R. J. & Ori-McKenney, K. M. Multiple modes of cytoplasmic dynein regulation. Nature Cell Biol.14, 224–230 (2012). ArticleCASPubMedGoogle Scholar
- Kardon, J. & Vale, R. Regulators of the cytoplasmic dynein motor. Nature Rev. Mol. Cell Biol.10, 854–865 (2009). ArticleCASGoogle Scholar
- Allan, V. J. Cytoplasmic dynein. Biochem. Soc. Trans.39, 1169–1178 (2011). ArticleCASPubMedGoogle Scholar
- Akhmanova, A. & Hammer, J. A. 3rd. Linking molecular motors to membrane cargo. Curr. Opin. Cell Biol.22, 479–487 (2010). ArticlePubMedPubMed CentralCASGoogle Scholar
- Riedel-Kruse, I. H., Hilfinger, A., Howard, J. & Jülicher, F. How molecular motors shape the flagellar beat. HFSP J.1, 192–208 (2007). ArticlePubMedPubMed CentralGoogle Scholar
- Brokaw, C. Thinking about flagellar oscillation. Cell. Motil. Cytoskeleton66, 425–436 (2009). ArticleCASPubMedGoogle Scholar
- Lindemann, C. B. & Lesich, K. A. Flagellar and ciliary beating: the proven and the possible. J. Cell Sci.123, 519–528 (2010). ArticleCASPubMedGoogle Scholar
- King, S. M. Integrated control of axonemal dynein AAA + motors. J. Struct. Biol.179, 222–228 (2012). ArticlePubMedPubMed CentralCASGoogle Scholar
- Wickstead, B. & Gull, K. Dyneins across eukaryotes: a comparative genomic analysis. Traffic8, 1708–1721 (2007). ArticlePubMedPubMed CentralCASGoogle Scholar
- Vale, R. D. The molecular motor toolbox for intracellular transport. Cell112, 467–480 (2003). ArticleCASPubMedGoogle Scholar
- Yagi, T. Bioinformatic approaches to dynein heavy chain classification. Methods Cell Biol.92, 1–9 (2009). ArticleCASPubMedGoogle Scholar
- Moore, J., Stuchell-Brereton, M. & Cooper, J. Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell. Motil. Cytoskeleton66, 546–555 (2009). ArticlePubMedPubMed CentralCASGoogle Scholar
- Egan, M. J., McClintock, M. A. & Reck-Peterson, S. L. Microtubule-based transport in filamentous fungi. Curr. Opin. Microbiol.15, 637–645 (2012). ArticlePubMedPubMed CentralCASGoogle Scholar
- Koonce, M. P. Dictyostelium, a model organism for microtubule-based transport. Protist151, 17–25 (2000). ArticleCASPubMedGoogle Scholar
- Pfister, K. K. et al. Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet.2, e1 (2006). ArticlePubMedPubMed CentralCASGoogle Scholar
- Trokter, M., Mücke, N. & Surrey, T. Reconstitution of the human cytoplasmic dynein complex. Proc. Natl Acad. Sci. USA109, 20895–20900 (2012). Describes the reconstitution of the human cytoplasmic dynein complex from recombinant subunits. Surprisingly, despite driving robust microtubule gliding, the complexes do not show processive motility, suggesting that additional factors might be required for this behaviour.ArticleCASPubMedPubMed CentralGoogle Scholar
- Schroer, T. A. Dynactin. Annu. Rev. Cell Dev. Biol.20, 759–779 (2004). ArticleCASPubMedGoogle Scholar
- Driskell, O. J., Mironov, A., Allan, V. J. & Woodman, P. G. Dynein is required for receptor sorting and the morphogenesis of early endosomes. Nature Cell Biol.9, 113–120 (2007). ArticleCASPubMedGoogle Scholar
- Jordens, I. et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol.11, 1680–1685 (2001). ArticleCASPubMedGoogle Scholar
- Blocker, A. et al. Molecular requirements for bi-directional movement of phagosomes along microtubules. J. Cell Biol.137, 113–129 (1997). ArticlePubMedPubMed CentralCASGoogle Scholar
- Gross, S. P. et al. Interactions and regulation of molecular motors in Xenopus melanophores. J. Cell Biol.156, 855–865 (2002). ArticlePubMedPubMed CentralCASGoogle Scholar
- Kural, C. et al. Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science308, 1469–1472 (2005). ArticleCASPubMedGoogle Scholar
- Gross, S. P., Welte, M. A., Block, S. M. & Wieschaus, E. F. Dynein-mediated cargo transport in vivo. A switch controls travel distance. J. Cell Biol.148, 945–956 (2000). ArticlePubMedPubMed CentralCASGoogle Scholar
- Pilling, A. D., Horiuchi, D., Lively, C. M. & Saxton, W. M. Kinesin-1 and dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell17, 2057–2068 (2006). ArticlePubMedPubMed CentralCASGoogle Scholar
- Presley, J. F. et al. ER-to-Golgi transport visualized in living cells. Nature389, 81–85 (1997). ArticleCASPubMedGoogle Scholar
- Heerssen, H. M., Pazyra, M. F. & Segal, R. A. Dynein motors transport activated Trks to promote survival of target-dependent neurons. Nature Neurosci.7, 596–604 (2004). ArticleCASPubMedGoogle Scholar
- Young, A., Dictenberg, J. B., Purohit, A., Tuft, R. & Doxsey, S. J. Cytoplasmic dynein-mediated assembly of pericentrin and gamma tubulin onto centrosomes. Mol. Biol. Cell11, 2047–2056 (2000). ArticlePubMedPubMed CentralCASGoogle Scholar
- Harrell, J. M. et al. Evidence for glucocorticoid receptor transport on microtubules by dynein. J. Biol. Chem.279, 54647–54654 (2004). ArticleCASPubMedGoogle Scholar
- Shah, J. V., Flanagan, L. A., Janmey, P. A. & Leterrier, J. F. Bidirectional translocation of neurofilaments along microtubules mediated in part by dynein/dynactin. Mol. Biol. Cell11, 3495–3508 (2000). ArticlePubMedPubMed CentralCASGoogle Scholar
- Wilkie, G. S. & Davis, I. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell105, 209–219 (2001). ArticleCASPubMedGoogle Scholar
- Johnston, J. A., Illing, M. E. & Kopito, R. R. Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell. Motil. Cytoskeleton53, 26–38 (2002). ArticleCASPubMedGoogle Scholar
- Maday, S., Wallace, K. E. & Holzbaur, E. L. F. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol.196, 407–417 (2012). ArticlePubMedPubMed CentralCASGoogle Scholar
- Dodding, M. P. & Way, M. Coupling viruses to dynein and kinesin-1. EMBO J.30, 3527–3539 (2011). ArticlePubMedPubMed CentralCASGoogle Scholar
- Sato, A. et al. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell139, 907–919 (2009). ArticlePubMedPubMed CentralCASGoogle Scholar
- Steinberg, G. et al. Motor-driven motility of fungal nuclear pores organizes chromosomes and fosters nucleocytoplasmic transport. J. Cell Biol.198, 343–355 (2012). ArticlePubMedPubMed CentralCASGoogle Scholar
- Schnyder, T. et al. B cell receptor-mediated antigen gathering requires ubiquitin ligase Cbl and adaptors Grb2 and Dok-3 to recruit dynein to the signaling microcluster. Immunity34, 905–918 (2011). ArticleCASPubMedGoogle Scholar
- Hashimoto-Tane, A. et al. Dynein-driven transport of T cell receptor microclusters regulates immune synapse formation and T cell activation. Immunity34, 919–931 (2011). ArticleCASPubMedGoogle Scholar
- Laan, L. et al. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell148, 502–514 (2012). ArticlePubMedPubMed CentralCASGoogle Scholar
- Hendricks, A. G. et al. Dynein tethers and stabilizes dynamic microtubule plus ends. Curr. Biol.22, 632–637 (2012). ArticlePubMedPubMed CentralCASGoogle Scholar
- Tsai, J.-W., Bremner, K. H. & Vallee, R. B. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nature Neurosci.10, 970–979 (2007). ArticleCASPubMedGoogle Scholar
- Dujardin, D. L. et al. A role for cytoplasmic dynein and LIS1 in directed cell movement. J. Cell Biol.163, 1205–1211 (2003). ArticlePubMedPubMed CentralCASGoogle Scholar
- Combs, J. et al. Recruitment of dynein to the Jurkat immunological synapse. Proc. Natl Acad. Sci. USA103, 14883–14888 (2006). ArticleCASPubMedPubMed CentralGoogle Scholar
- McNally, F. J. Mechanisms of spindle positioning. J. Cell Biol.200, 131–140 (2013). ArticlePubMedPubMed CentralCASGoogle Scholar
- Kiyomitsu, T. & Cheeseman, I. M. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nature Cell Biol.14, 311–317 (2012). ArticleCASPubMedGoogle Scholar
- Collins, E. S., Balchand, S. K., Faraci, J. L., Wadsworth, P. & Lee, W.-L. Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol. Biol. Cell23, 3380–3390 (2012). ArticlePubMedPubMed CentralCASGoogle Scholar
- Mitchison, T. et al. Growth, interaction, and positioning of microtubule asters in extremely large vertebrate embryo cells. Cytoskeleton (Hoboken)69, 738–750 (2012). ArticleCASGoogle Scholar
- Kimura, K. & Kimura, A. Intracellular organelles mediate cytoplasmic pulling force for centrosome centration in the Caenorhabditis elegans early embryo. Proc. Natl Acad. Sci. USA108, 137–142 (2011). ArticleCASPubMedGoogle Scholar
- Longoria, R. A. & Shubeita, G. T. Cargo transport by cytoplasmic dynein can center embryonic centrosomes. PLoS ONE8, e67710 (2013). ArticlePubMedPubMed CentralCASGoogle Scholar
- Corthesy-Theulaz, I., Pauloin, A. & Pfeffer, S. R. Cytoplasmic dynein participates in the centrosomal localization of the Golgi complex. J. Cell Biol.118, 1333–1345 (1992). ArticleCASPubMedGoogle Scholar
- Levy, J. R. & Holzbaur, E. L. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J. Cell Sci.121, 3187–3195 (2008). ArticlePubMedCASGoogle Scholar
- Raaijmakers, J. A. et al. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J.31, 4179–4190 (2012). ArticlePubMedPubMed CentralCASGoogle Scholar
- Salina, D. et al. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell108, 97–107 (2002). ArticleCASPubMedGoogle Scholar
- Heald, R. et al. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature382, 420–425 (1996). ArticleCASPubMedGoogle Scholar
- Merdes, A., Ramyar, K., Vechio, J. D. & Cleveland, D. W. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell87, 447–458 (1996). ArticleCASPubMedGoogle Scholar
- Foley, E. A. & Kapoor, T. M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nature Rev. Mol. Cell Biol.14, 25–37 (2012). ArticleCASGoogle Scholar
- Howell, B. J. et al. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol.155, 1159–1172 (2001). ArticlePubMedPubMed CentralCASGoogle Scholar
- Wojcik, E. et al. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nature Cell Biol.3, 1001–1007 (2001). ArticleCASPubMedGoogle Scholar
- Varma, D., Monzo, P., Stehman, S. A. & Vallee, R. B. Direct role of dynein motor in stable kinetochore–microtubule attachment, orientation, and alignment. J. Cell Biol.182, 1045–1054 (2008). ArticlePubMedPubMed CentralCASGoogle Scholar
- Yang, Z., Tulu, U. S., Wadsworth, P. & Rieder, C. L. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr. Biol.17, 973–980 (2007). ArticlePubMedPubMed CentralCASGoogle Scholar
- Ishikawa, H. & Marshall, W. F. Ciliogenesis: building the cell's antenna. Nature Rev. Mol. Cell Biol.12, 222–234 (2011). ArticleCASGoogle Scholar
- Pazour, G. J., Dickert, B. L. & Witman, G. B. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol.144, 473–481 (1999). ArticlePubMedPubMed CentralCASGoogle Scholar
- Porter, M. E., Bower, R., Knott, J. A., Byrd, P. & Dentler, W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell10, 693–712 (1999). ArticlePubMedPubMed CentralCASGoogle Scholar
- Mikami, A. et al. Molecular structure of cytoplasmic dynein 2 and its distribution in neuronal and ciliated cells. J. Cell Sci.115, 4801–4808 (2002). ArticleCASPubMedGoogle Scholar
- Perrone, C. A. et al. A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in Chlamydomonas and mammalian cells. Mol. Biol. Cell14, 2041–2056 (2003). ArticlePubMedPubMed CentralCASGoogle Scholar
- Ichikawa, M., Watanabe, Y., Murayama, T. & Toyoshima, Y. Y. Recombinant human cytoplasmic dynein heavy chain 1 and 2: observation of dynein-2 motor activity in vitro. FEBS Lett.585, 2419–2423 (2011). ArticleCASPubMedGoogle Scholar
- Pigino, G. et al. Electron-tomographic analysis of intraflagellar transport particle trains in situ. J. Cell Biol.187, 135–148 (2009). ArticlePubMedPubMed CentralCASGoogle Scholar
- Laib, J. A., Marin, J. A., Bloodgood, R. A. & Guilford, W. H. The reciprocal coordination and mechanics of molecular motors in living cells. Proc. Natl Acad. Sci. USA106, 3190–3195 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Shih, S. M. et al. Intraflagellar transport drives flagellar surface motility. Elife2, e00744 (2013). ArticlePubMedPubMed CentralGoogle Scholar
- Mallik, R., Rai, A. K., Barak, P., Rai, A. & Kunwar, A. Teamwork in microtubule motors. Trends Cell Biol.http://dx.doi.org/10.1016/j.tcb.2013.06.003 (2013).
- Koonce, M. P. & Samsó, M. Overexpression of cytoplasmic dynein's globular head causes a collapse of the interphase microtubule network in Dictyostelium. Mol. Biol. Cell7, 935–948 (1996). Details a recombinant expression system for theD. discoideumcytoplasmic dynein motor domain, forming the foundation for numerous structural and functional analyses. Also demonstrates that the motor domain includes the globular head structure seen in earlier electron microscopy images.ArticlePubMedPubMed CentralCASGoogle Scholar
- Nishiura, M. et al. A single-headed recombinant fragment of Dictyostelium cytoplasmic dynein can drive the robust sliding of microtubules. J. Biol. Chem.279, 22799–22802 (2004). ArticleCASPubMedGoogle Scholar
- Reck-Peterson, S. L. et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell126, 335–348 (2006). EstablishesS. cerevisiaeas another important cytoplasmic dynein expression system and provides insight into the requirements for processive dynein motion.ArticlePubMedPubMed CentralCASGoogle Scholar
- Höök, P. et al. Long range allosteric control of cytoplasmic dynein ATPase activity by the stalk and C-terminal domains. J. Biol. Chem.280, 33045–33054 (2005). ArticleCASPubMedGoogle Scholar
- Imamula, K., Kon, T., Ohkura, R. & Sutoh, K. The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. Proc. Natl Acad. Sci. USA104, 16134–16139 (2007). ArticleCASPubMedPubMed CentralGoogle Scholar
- Mogami, T., Kon, T., Ito, K. & Sutoh, K. Kinetic characterization of tail swing steps in the ATPase cycle of Dictyostelium cytoplasmic dynein. J. Biol. Chem.282, 21639–21644 (2007). ArticleCASPubMedGoogle Scholar
- Johnson, K. A. Pathway of the microtubule–dynein ATPase and the structure of dynein: a comparison with actomyosin. Annu. Rev. Biophys. Biophys. Chem.14, 161–188 (1985). ArticleCASPubMedGoogle Scholar
- Samsó, M., Radermacher, M., Frank, J. & Koonce, M. P. Structural characterization of a dynein motor domain. J. Mol. Biol.276, 927–937 (1998). ArticlePubMedGoogle Scholar
- Gee, M. A., Heuser, J. E. & Vallee, R. B. An extended microtubule-binding structure within the dynein motor domain. Nature390, 636–639 (1997). ArticleCASPubMedGoogle Scholar
- Roberts, A. J. et al. ATP-driven remodeling of the linker domain in the dynein motor. Structure20, 1670–1680 (2012). A 2D and 3D electron microscopy study of an axonemal and cytoplasmic dynein, revealing that the linker domain is a stable structural entity that is remodelled by the AAA+ ring.ArticlePubMedPubMed CentralCASGoogle Scholar
- Roberts, A. et al. AAA+ ring and linker swing mechanism in the dynein motor. Cell136, 485–495 (2009). ArticlePubMedPubMed CentralCASGoogle Scholar
- Burgess, S. A., Walker, M. L., Sakakibara, H., Knight, P. J. & Oiwa, K. Dynein structure and power stroke. Nature421, 715–718 (2003). ArticleCASPubMedGoogle Scholar
- Kon, T. et al. The 2.8 Å crystal structure of the dynein motor domain. Nature484, 345–350 (2012). Reports a high-resolution crystal structure of theD. discoideumdynein motor domain in the ADP-bound state. Structure-guided mutagenesis provides insight into how the linker domain moves and the roles of the AAA+ modules, strut and C-terminal sequence in allosteric communication.ArticleCASPubMedGoogle Scholar
- Kon, T., Sutoh, K. & Kurisu, G. X-ray structure of a functional full-length dynein motor domain. Nature Struct. Mol. Biol.18, 638–642 (2011). ArticleCASGoogle Scholar
- Schmidt, H., Gleave, E. S. & Carter, A. P. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nature Struct. Mol. Biol.19, 492–497 (2012). Reports a high-resolution structure of theS. cerevisiaedynein motor domain, crystallized in the absence of nucleotide. Crystal-soaking experiments reveal distinct nucleotide-binding behaviours in AAA1–AAA4, and mutagenesis suggests a functional role for linker docking at AAA5.ArticleCASGoogle Scholar
- Carter, A. P., Cho, C., Jin, L. & Vale, R. D. Crystal structure of the dynein motor domain. Science331, 1159–1165 (2011). ArticlePubMedPubMed CentralCASGoogle Scholar
- Glynn, S. E., Martin, A., Nager, A. R., Baker, T. A. & Sauer, R. T. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell139, 744–756 (2009). ArticlePubMedPubMed CentralCASGoogle Scholar
- Wang, J. et al. Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure9, 1107–1116 (2001). ArticleCASPubMedGoogle Scholar
- Gibbons, I. R. et al. Photosensitized cleavage of dynein heavy chains. Cleavage at the “V1 site” by irradiation at 365 nm in the presence of ATP and vanadate. J. Biol. Chem.262, 2780–2786 (1987). CASPubMedGoogle Scholar
- Kon, T., Nishiura, M., Ohkura, R., Toyoshima, Y. Y. & Sutoh, K. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry43, 11266–11274 (2004). ArticleCASPubMedGoogle Scholar
- Enemark, E. J. & Joshua-Tor, L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature442, 270–275 (2006). ArticleCASPubMedGoogle Scholar
- Cho, C., Reck-Peterson, S. & Vale, R. Cytoplasmic dynein's regulatory ATPase sites affect processivity and force generation. J. Biol. Chem.283, 25839–25845 (2008). ArticlePubMedPubMed CentralCASGoogle Scholar
- Huang, J., Roberts, A. J., Leschziner, A. E. & Reck-Peterson, S. L. Lis1 acts as a “clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell150, 975–986 (2012). ArticlePubMedPubMed CentralCASGoogle Scholar
- McKenney, R. J., Vershinin, M., Kunwar, A., Vallee, R. B. & Gross, S. P. LIS1 and NudE induce a persistent dynein force-producing state. Cell141, 304–314 (2010). ArticlePubMedPubMed CentralCASGoogle Scholar
- Yamada, M. et al. LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J.27, 2471–2483 (2008). ArticlePubMedPubMed CentralCASGoogle Scholar
- Cho, C. & Vale, R. D. The mechanism of dynein motility: insight from crystal structures of the motor domain. Biochim. Biophys. Acta1823, 182–191 (2011). ArticlePubMedPubMed CentralCASGoogle Scholar
- Numata, N., Shima, T., Ohkura, R., Kon, T. & Sutoh, K. C-Sequence of the Dictyostelium cytoplasmic dynein participates in processivity modulation. FEBS Lett.585, 1185–1190 (2011). ArticleCASPubMedGoogle Scholar
- Hwang, W. & Lang, M. J. Nucleotide-dependent control of internal strains in ring-shaped AAA+ motors. Cell. Mol. Bioeng.6, 65–73 (2013). ArticleCASPubMedGoogle Scholar
- Gibbons, I. R. et al. The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. J. Biol. Chem.280, 23960–23965 (2005). ArticlePubMedCASGoogle Scholar
- Carter, A. P. et al. Structure and functional role of dynein's microtubule-binding domain. Science322, 1691–1695 (2008). ArticlePubMedPubMed CentralCASGoogle Scholar
- Kon, T. et al. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nature Struct. Mol. Biol.16, 325–333 (2009). ArticleCASGoogle Scholar
- Redwine, W. B. et al. Structural basis for microtubule binding and release by dynein. Science337, 1532–1536 (2012). References 109–112 provide evidence for the helix-sliding hypothesis for allosteric communication through the stalk of dynein, which was put forward in reference 109. Reference 112 also provides insight into the structural changes within the microtubule-binding domain of dynein.ArticlePubMedPubMed CentralCASGoogle Scholar
- Sweeney, H. L. & Houdusse, A. The motor mechanism of myosin V: insights for muscle contraction. Philo. Trans. R. Soc. Lond. B359, 1829–1841 (2004). ArticleCASGoogle Scholar
- Xi, Z., Gao, Y., Sirinakis, G., Guo, H. & Zhang, Y. Single-molecule observation of helix staggering, sliding, and coiled coil misfolding. Proc. Natl Acad. Sci. USA109, 5711–5716 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Burgess, S. A., Walker, M. L., Sakakibara, H., Oiwa, K. & Knight, P. J. The structure of dynein-c by negative stain electron microscopy. J. Struct. Biol.146, 205–216 (2004). ArticleCASPubMedGoogle Scholar
- Burgess, S. A. & Knight, P. J. Is the dynein motor a winch? Curr. Opin. Struct. Biol.14, 138–146 (2004). ArticleCASPubMedGoogle Scholar
- Gennerich, A., Carter, A. P., Reck-Peterson, S. L. & Vale, R. D. Force-induced bidirectional stepping of cytoplasmic dynein. Cell131, 952–965 (2007). ArticlePubMedPubMed CentralCASGoogle Scholar
- Kon, T., Mogami, T., Ohkura, R., Nishiura, M. & Sutoh, K. ATP hydrolysis cycle-dependent tail motions in cytoplasmic dynein. Nature Struct. Mol. Biol.12, 513–519 (2005). ArticleCASGoogle Scholar
- Shima, T., Kon, T., Imamula, K., Ohkura, R. & Sutoh, K. Two modes of microtubule sliding driven by cytoplasmic dynein. Proc. Natl Acad. Sci. USA103, 17736–17740 (2006). ArticleCASPubMedPubMed CentralGoogle Scholar
- Ueno, H., Yasunaga, T., Shingyoji, C. & Hirose, K. Dynein pulls microtubules without rotating its stalk. Proc. Natl Acad. Sci. USA105, 19702–19707 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- Movassagh, T., Bui, K. H., Sakakibara, H., Oiwa, K. & Ishikawa, T. Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis. Nature Struct. Mol. Biol.17, 761–767 (2010). ArticleCASGoogle Scholar
- Qiu, W. et al. Dynein achieves processive motion using both stochastic and coordinated stepping. Nature Struct. Mol. Biol.19, 193–200 (2012). ArticleCASGoogle Scholar
- Dewitt, M. A., Chang, A. Y., Combs, P. A. & Yildiz, A. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science335, 221–225 (2011). References 122 and 123 use two-colour single-molecule experiments to reveal thatS. cerevisiaecytoplasmic dynein dimers can move processively along the microtubule without following a strict stepping pattern. Analysis of the stepping traces provides insight into the spatial configuration of the motor domains and their loose, tension-based coordination.ArticlePubMedPubMed CentralCASGoogle Scholar
- Veigel, C., Wang, F., Bartoo, M. L., Sellers, J. R. & Molloy, J. E. The gated gait of the processive molecular motor, myosin V. Nature Cell Biol.4, 59–65 (2002). ArticleCASPubMedGoogle Scholar
- Shima, T., Imamula, K., Kon, T., Ohkura, R. & Sutoh, K. Head-head coordination is required for the processive motion of cytoplasmic dynein, an AAA+ molecular motor. J. Struct. Biol.156, 182–189 (2006). ArticleCASPubMedGoogle Scholar
- Stinson, B. M. et al. Nucleotide binding and conformational switching in the hexameric ring of a AAA+ machine. Cell153, 628–639 (2013). ArticlePubMedPubMed CentralCASGoogle Scholar
- Sakamoto, T., Webb, M. R., Forgacs, E., White, H. D. & Sellers, J. R. Direct observation of the mechanochemical coupling in myosin Va during processive movement. Nature455, 128–132 (2008). ArticlePubMedPubMed CentralCASGoogle Scholar
- Firestone, A. J. et al. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature484, 125–129 (2012). ArticlePubMedPubMed CentralCASGoogle Scholar
- Kamiya, R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int. Rev. Cytol.219, 115–155 (2002). ArticleCASPubMedGoogle Scholar
- Kikushima, K. & Kamiya, R. Clockwise translocation of microtubules by flagellar inner-arm dyneins in vitro. Biophys. J.94, 4014–4019 (2008). ArticlePubMedPubMed CentralCASGoogle Scholar
- Mallik, R., Carter, B. C., Lex, S. A., King, S. J. & Gross, S. P. Cytoplasmic dynein functions as a gear in response to load. Nature427, 649–652 (2004). ArticleCASPubMedGoogle Scholar
- Rai, A. K., Rai, A., Ramaiya, A. J., Jha, R. & Mallik, R. Molecular adaptations allow dynein to generate large collective forces inside cells. Cell152, 172–182 (2013). ArticleCASPubMedGoogle Scholar
- Schroeder, H. W., Mitchell, C., Shuman, H., Holzbaur, E. L. F. & Goldman, Y. E. Motor number controls cargo switching at actin-microtubule intersections in vitro. Curr. Biol.20, 687–696 (2010). ArticleCASPubMedGoogle Scholar
- Toba, S., Watanabe, T. M., Yamaguchi-Okimoto, L., Toyoshima, Y. Y. & Higuchi, H. Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proc. Natl Acad. Sci. USA103, 5741–5745 (2006). ArticleCASPubMedPubMed CentralGoogle Scholar
- Walter, W. J., Koonce, M. P., Brenner, B. & Steffen, W. Two independent switches regulate cytoplasmic dynein's processivity and directionality. Proc. Natl Acad. Sci. USA109, 5289–5293 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Ross, J. L., Wallace, K., Shuman, H., Goldman, Y. E. & Holzbaur, E. L. F. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nature Cell Biol.8, 562–570 (2006). ArticleCASPubMedGoogle Scholar
- Ananthanarayanan, V. et al. Dynein motion switches from diffusive to directed upon cortical anchoring. Cell153, 1526–1536 (2013). ArticleCASPubMedGoogle Scholar
- Nicastro, D. et al. Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc. Natl Acad. Sci. USA108, E845–E853 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Summers, K. E. & Gibbons, I. R. Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc. Natl Acad. Sci. USA68, 3092–3096 (1971). ArticleCASPubMedPubMed CentralGoogle Scholar
- Nicastro, D. Cryo-electron microscope tomography to study axonemal organization. Methods Cell Biol.91, 1–39 (2009). ArticleCASPubMedGoogle Scholar
- Hom, E. F. et al. A unified taxonomy for ciliary dyneins. Cytoskeleton (Hoboken)68, 555–565 (2011). ArticleCASGoogle Scholar
- Bui, K. H., Yagi, T., Yamamoto, R., Kamiya, R. & Ishikawa, T. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J. Cell Biol.198, 913–925 (2012). ArticlePubMedPubMed CentralCASGoogle Scholar
- Kagami, O. & Kamiya, R. Translocation and rotation of microtubules caused by multiple species of Chlamydomonas inner-arm dynein. J. Cell Sci.103, 653–664 (1992). CASGoogle Scholar
- Yagi, T., Uematsu, K., Liu, Z. & Kamiya, R. Identification of dyneins that localize exclusively to the proximal portion of Chlamydomonas flagella. J. Cell Sci.122, 1306–1314 (2009). ArticleCASPubMedGoogle Scholar
- Woolley, D. M. Studies on the eel sperm flagellum. I. The structure of the inner dynein arm complex. J. Cell Sci.110, 85–94 (1997). CASPubMedGoogle Scholar
- Zhang, X. & Wigley, D. The 'glutamate switch' provides a link between ATPase activity and ligand binding in AAA+ proteins. Nature Struct. Mol. Biol.15, 1223–1227 (2008). ArticleCASGoogle Scholar
- Hanson, P. I. & Whiteheart, S. W. AAA+ proteins: have engine, will work. Nature Rev. Mol. Cell Biol.6, 519–529 (2005). ArticleCASGoogle Scholar
- Pfister, K. K. et al. Cytoplasmic dynein nomenclature. J. Cell Biol.171, 411–413 (2005). ArticlePubMedPubMed CentralCASGoogle Scholar
- Mizuno, N. et al. Dynein and kinesin share an overlapping microtubule-binding site. EMBO J.23, 2459–2467 (2004). ArticlePubMedPubMed CentralCASGoogle Scholar
Acknowledgements
The authors apologize to their colleagues whose work could not be cited owing to space limitations. They thank K. Toropova, R. Hernandez-Lopez, J. Huang and S. Reck-Peterson for helpful comments on the manuscript and B. Malkova for providing electron microscopy data for figure 5a. A.J.R. is grateful to J. Iwasa for training on AutoDesk Maya software. The work in the authors laboratory was supported by: a Sir Henry Wellcome Postdoctoral Fellowship (092436/Z/10/Z) to A.J.R.; a Grant-in-Aid for Scientific Research (B) 23370073 from the Japan Society for Promotion of Science (JSPS) and a Japan Science and Technology Agency PRESTO award to T.K.; a Grant-in-Aid for Scientific Research (B) 23370075 from the JSPS to K.S., and grants BB/E00928X/1 and BB/BB/K000705/1 from the BBSRC (UK) and RGP0009/2008-C from the Human Frontiers Science Program to S.A.B.
Author information
- S.A.Burgess@leeds.ac.uk
Authors and Affiliations
- Astbury Centre for Structural Molecular Biology, School of Molecular and Cellular Biology, Faculty of Biological Sciences, University of Leeds, Leeds, LS2 9JT, UK Anthony J. Roberts, Peter J. Knight & Stan A. Burgess
- Department of Cell Biology, Harvard Medical School, 240 Longwood Avenue, Boston, 02115, Massachusetts, USA Anthony J. Roberts
- Department of Frontier Bioscience, Faculty of Bioscience and Applied Chemistry, Hosei University, 3-7-2 Kajino-cho, Koganei, Tokyo, 184-8584, Japan Takahide Kon
- Japan Science and Technology Agency, PRESTO, 4-1-8 Honcho, Kawaguchi, 332-0012, Saitama, Japan Takahide Kon
- Faculty of Science and Engineering, Waseda University, Okubo 3-4-1, Shinjuku-ku, 169-8555, Tokyo, Japan Kazuo Sutoh
- Anthony J. Roberts









